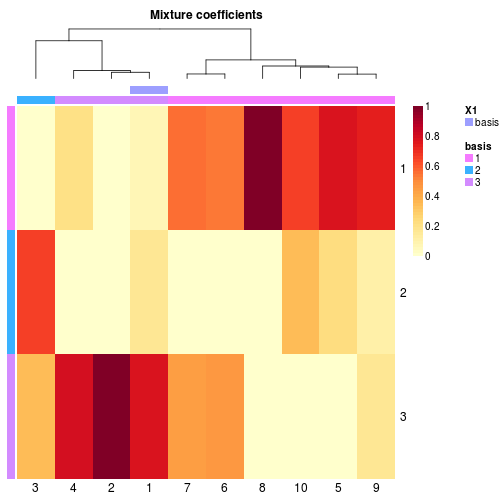
#' # random data with underlying NMF model
v <- syntheticNMF(20, 3, 10)
# estimate a model
x <- nmf(v, 3)
# highligh row only (using custom colors)
basismap(x, tracks=':basis', annColor=list(basis=1:3))
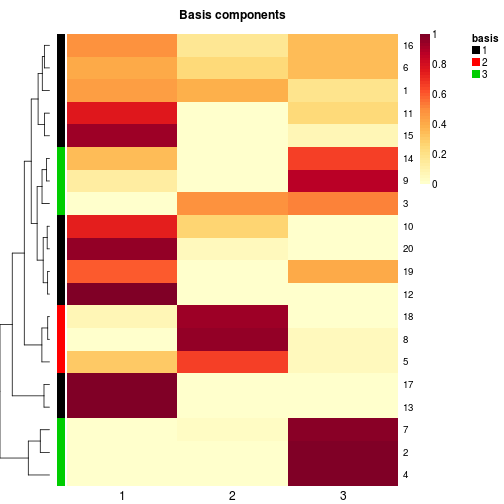
## character annotation vector: ok if it does not contain 'basis'
# annotate first and second row + automatic special track
basismap(x, annRow=c('alpha', 'beta'))
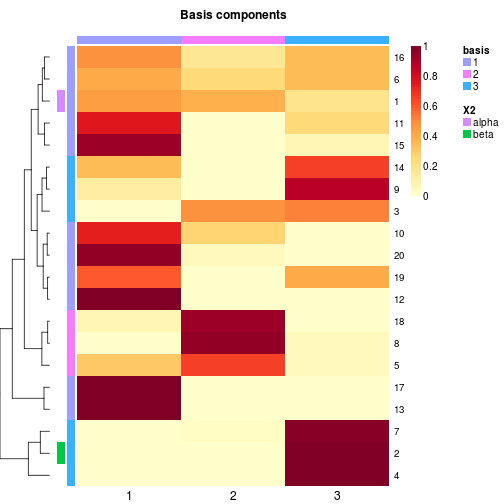
# no special track here
basismap(x, annRow=c('alpha', 'beta', ':basis'), tracks=NA)
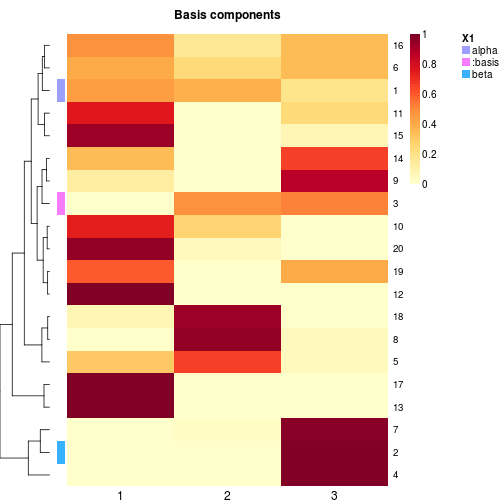
# with special track `basis`
basismap(x, annRow=list(c('alpha', 'beta'), ':basis'), tracks=NA)
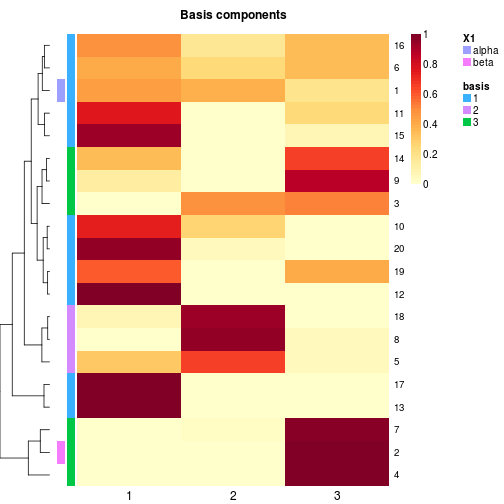
# highligh columns only (using custom colors)
basismap(x, tracks='basis:')
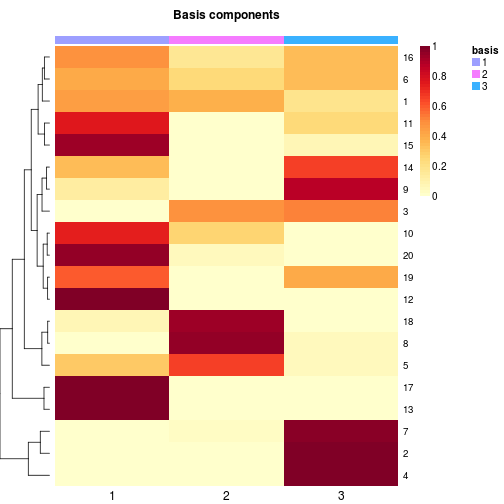
# changing the name of the basis annotation track
basismap(x, annRow=list(new_name=':basis'))
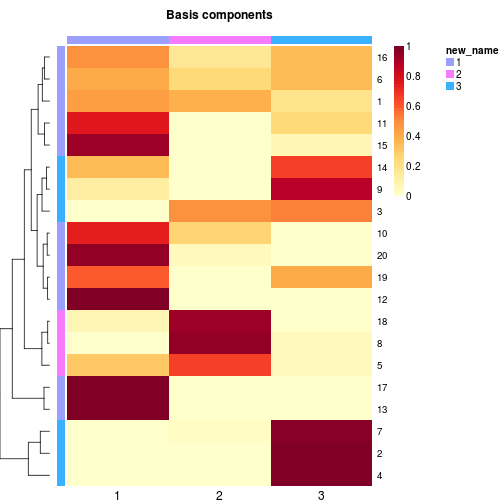
# coefficient matrix
coefmap(x, annCol=c('alpha', 'beta')) # annotate first and second sample
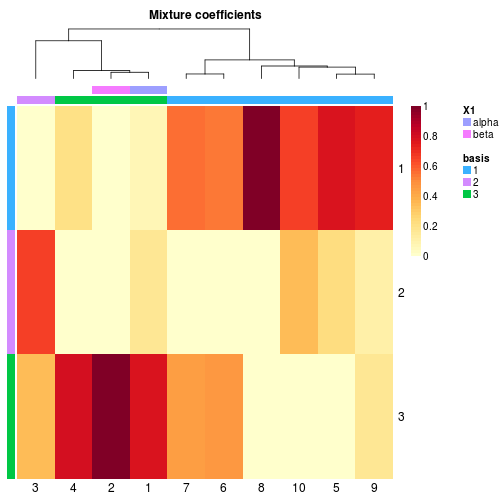
coefmap(x, annCol=list('basis', Greek=c('alpha', 'beta'))) # annotate first and second sample + basis annotation
coefmap(x, annCol=c(new_name='basis'))